

In addition, end-point measurement enables nucleic acid quantitation independently of the reaction efficiency, resulting in a positive-negative call for every droplet and greater amenability to multiplexed detection of target molecules 14. These factors offer the advantage of direct and independent quantification of DNA without standard curves giving more precise and reproducible data versus qPCR especially in the presence of sample contaminants that can partially inhibit Taq polymerase and/or primer annealing 11, 12, 13. However, there are two distinct differences: 1) the partitioning of the PCR reaction into thousands of individual reaction vessels prior to amplification and 2) the acquisition of data at reaction end point. As with qPCR, ddPCR technology utilizes Taq polymerase in a standard PCR reaction to amplify a target DNA fragment from a complex sample using pre-validated primer or primer/probe assays. The consequence of ignoring MIQE-guided protocols has led to the retraction of multiple articles over the past several years and remains a major frustration in the scientific community 8.ĭroplet Digital PCR (ddPCR) is a recent technology that has become commercially available since 2011 9, 10. The Minimum Information for the Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines and related articles published thereafter define a rigorous methodology for designing qPCR experiments that assures publication of reproducible and high quality data 5, 6, 7. For absolute quantification, data analysis is further complicated by the different sources of DNA from which the samples and standard curves are derived with unique backgrounds and contaminants that can variably affect the activity of Taq polymerase giving misleading results 4. Poorly optimized reactions can result in artifactual Cq values and misinterpreted data that are difficult or even impossible to reproduce 2, 3.
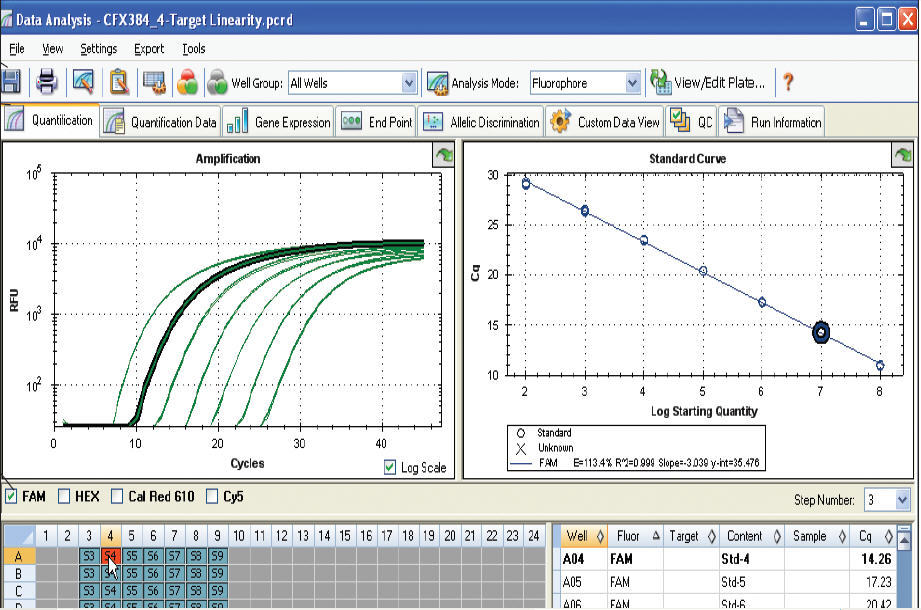
Therefore, optimization is critical for each primer pair such that reaction efficiency is consistent between all samples and acceptable (between 90% to 110%) with sample contaminants diluted adequately to assure that all reactions and associated Cq values are within the efficient range of the respective standard curves 1. A stepwise methodology is also described to choose between these complimentary technologies to obtain the best results for any experiment.ĭata from qPCR experiments are taken within each enzymatic reaction curve at the quantification cycle (Cq).
We conclude that for sample/target combinations with low levels of nucleic acids (Cq ≥ 29) and/or variable amounts of chemical and protein contaminants, ddPCR technology will produce more precise, reproducible and statistically significant results required for publication quality data. Here, Droplet Digital PCR (ddPCR) and qPCR platforms were directly compared for gene expression analysis using low amounts of purified, synthetic DNA in well characterized samples under identical reaction conditions. The most susceptible, frustrating and often most interesting samples are those containing low abundant targets with small expression differences of 2-fold or lower. The root cause of poor quality data is typically associated with inadequate dilution of residual protein and chemical contaminants that variably inhibit Taq polymerase and primer annealing.
#BIO RAD CFX MANAGER MELT CURVE VERIFICATION#
Quantitative PCR (qPCR) has become the gold standard technique to measure cDNA and gDNA levels but the resulting data can be highly variable, artifactual and non-reproducible without appropriate verification and validation of both samples and primers.


 0 kommentar(er)
0 kommentar(er)
